Accumulating evidence about the interaction between human microbiota and therapeutic-related substances, especially for clinical used drugs, deepens and broadens our understanding in the role of "our second genome" in human health and diseases, and opens a new window for managing/treating human healthy problems/diseases by manipulating microbiota-human interactions.
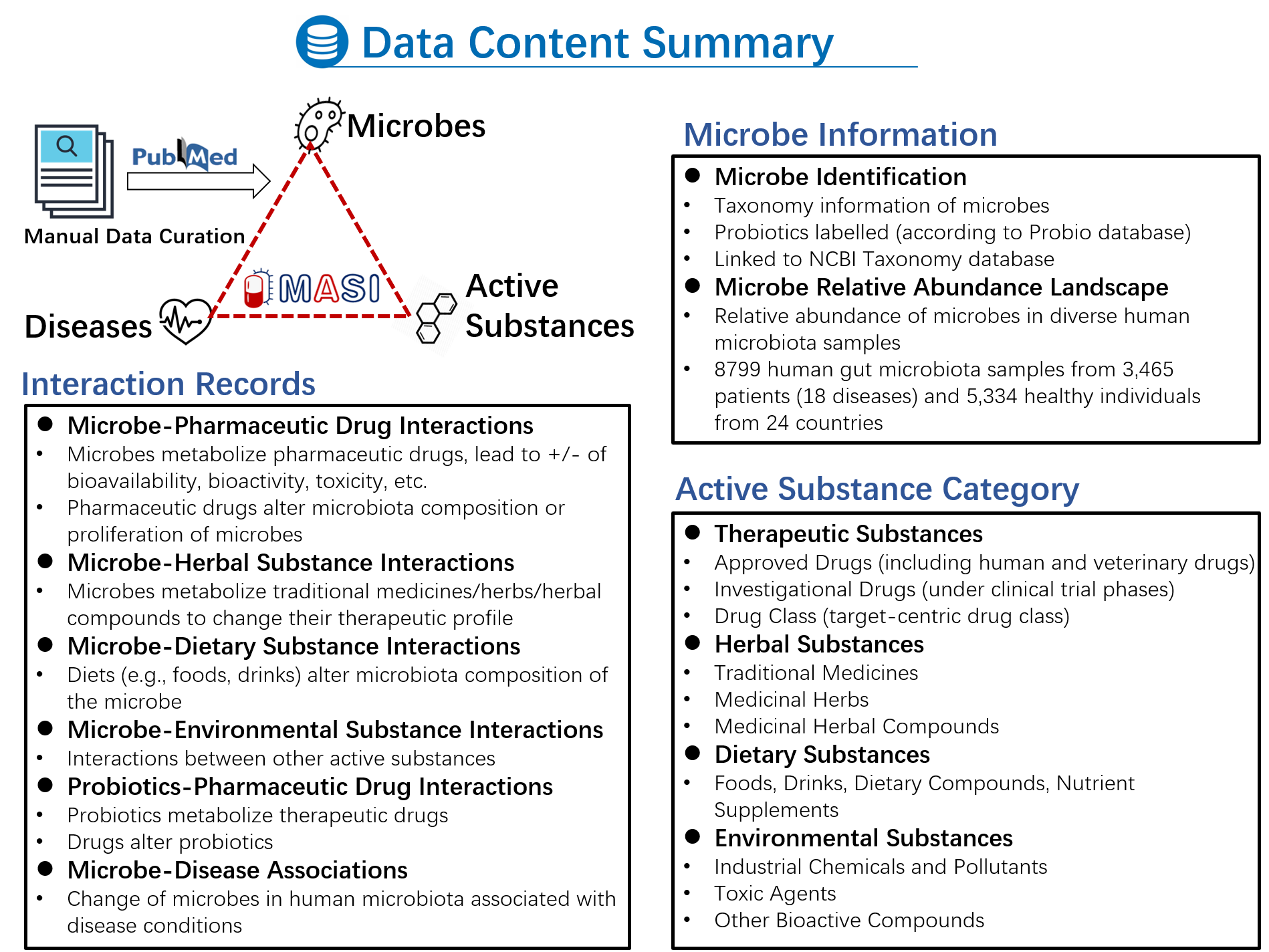
A centralized and standardized database to catalog Microbiota-Active Substances Interactions (MASIs) will be very useful for related research fields, including microbiota-related pharmaceutical research, food and nutrition research, clinical pharmacy, health management studies. Here, the MASI database is developed to cater this urgent need by providing manually curated MASIs data from scientific literature.
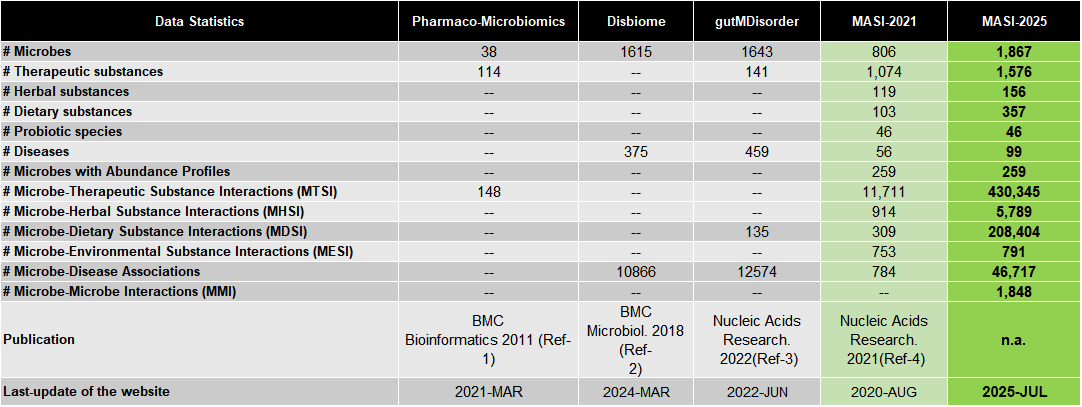
In the recent update to MASI v2.0, we have significantly expanded the scale of data and reorganized our database to meet recent trends:
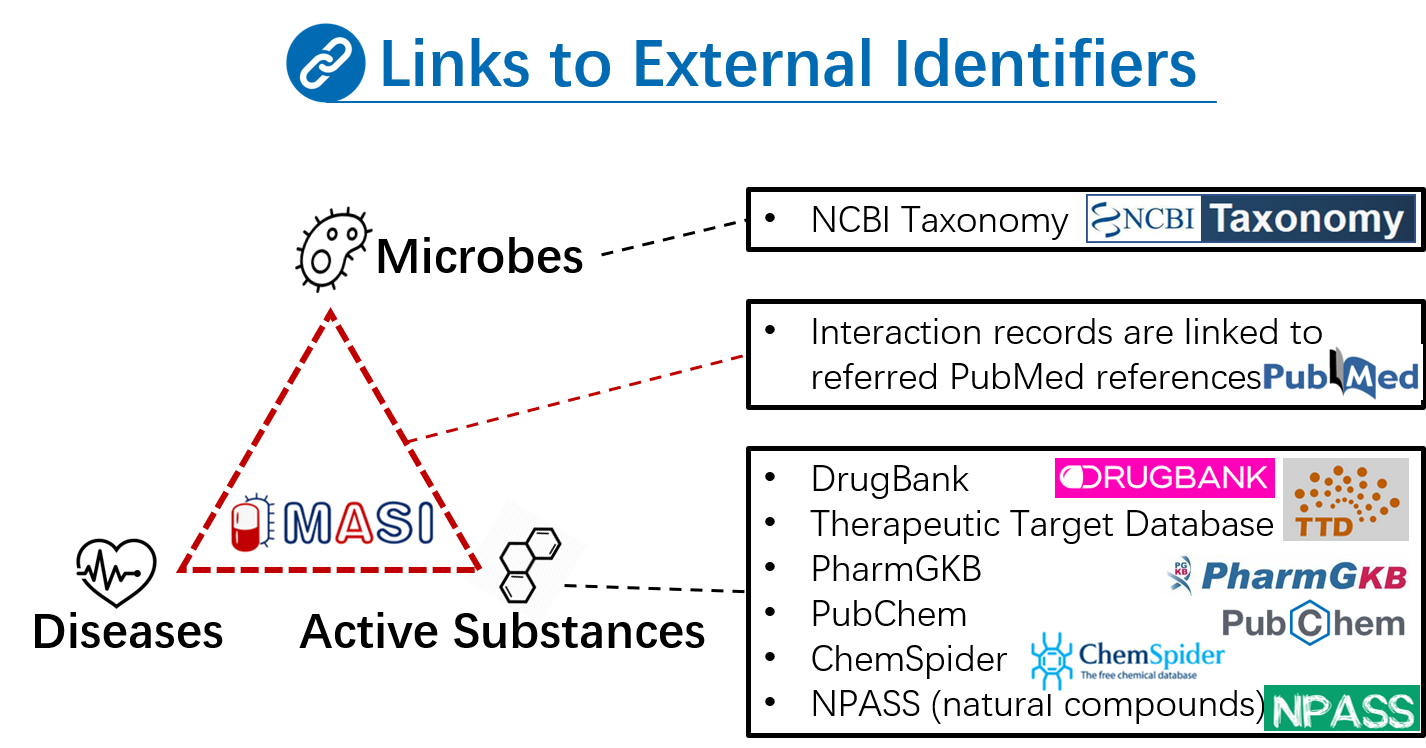
To better support this reorganization, in MASI v2.0, we implemented multi-source integration at the strain level and developed a comprehensive platform to characterize microbes using various types of evidence, making it a one-stop database for users. We have integrated several new categories of data, which are detailed as follows:
To elucidate microbes' potential capacities for metabolizing specific substrates and biosynthesizing secondary metabolites, we firstly collected 1,171 species' reference genome from RefSeq. Then two tools for identifying potential gene clusters were used (antiSMASH [Ref: Nucleic Acids Research, 51.W1 (2023), W46–W50] and gutSMASH [Ref: Nucleic Acids Research, 49.W1 (2021), W263–W270] ) to process reference genomes. Next, we manually examined results and collected supportive publications and information in the external databases to connect microbes with the substances they may metabolize or synthesize. Totally, 2449 known BGCs and 379 know MGCs with 100% similarity were found, linking to 648 related primary or secondary metabolites.
We collected genomes from reported publications and followed the pipeline discribed in AGORA2 [Ref: Nature Biotechnology, 41 (2023) 1320?331] and AGREDA [Ref:Nature Communications, 12 (2021) 4728] By extracting drug- and dietary-related metabolic genes, together with the relevant substrates, we matched the corresponding chemical reactions with microbes and linked them with the corresponding substrates. Finally, we categorized the reactions by their reconstruction subsystems and visualized all related substances and metabolites for each microbe. This brought in new 131 drugs and 366 dietary substances. Totally 1,781,579 records of microbe-therapeutic-substance records and 257,690 records of microbe-dietary-substance records were added (including different chemical forms).
Quorum sensing is a communication mechanism utilized by microbes, with specific molecules acting as QS languages. We collected strain- and species-level QS language records by examining publications, databases and recent machine learning methods (including QSHGM Database [Ref: Nat Commun 13, 3079 (2022)] and QSDB Database [Ref: Database (Oxford). 2021 Nov 26;2021(2021):baab071] databases etc.), displaying each microbe-QS-language pair, merging all languages at the species level, and linking all species to their corresponding QS languages. This generated a comprehensive interactive map indicating potential interactions. In total, 36 different QS languages covering 1848 records of microbe-QS-language pairs were added.
To illustrate microbes' distribution and their impact on different human organs, we collected data from recent publications and existing databases, categorizing them by different body sites and visualized their distribution, abundance, and association with human diseases via Image Map.
To add more recent information to our original database, we collected additional data from published articles and external databases. A total of 1,679 addtional articles were added to strengthen our database.
We employed more efficient visualization methods to display our data and evaluated the information abundance for each data category, selecting the most informative microbes to feature on our browse page.
Ref-1. Ramy K Aziz, Rama Saad, and Mariam R Rizkallah. PharmacoMicrobiomics or how bugs modulate drugs: an educational initiative to explore the effects of human microbiome on drugs. BMC Bioinformatics 2011; 12(Suppl 7): A10. Resource URL: http://www.pharmacomicrobiomics.com
Ref-2. Yorick Janssens, Joachim Nielandt, Antoon Bronselaer, et al. Disbiome Database: Linking the Microbiome to Disease. BMC Microbiol. 2018; 18(1):50. Resource URL: https://disbiome.ugent.be/home
Ref-3. Liang Cheng, Changlu Qi, He Zhuang, et al. gutMDisorder: A Comprehensive Database for Dysbiosis of the Gut Microbiota in Disorders and Interventions. Nucleic Acids Res 2020; 48(D1):D554-D560. Resource URL: http://bio-annotation.cn/gutMDisorder/home.dhtml
Ref-4. Zeng X, Yang X, Fan J, et al. MASI: microbiota—active substance interactions database[J]. Nucleic Acids Res 2021; 49(D1): D776-D782. http://www.aiddlab.com/MASI2025/index.html